 |
|
News and Publications
Synthesis and 3-Dimensional Visualization of Multiresolution Structural Data
W. Wriggers, P. Chacón, J. Kovacs, S. Birmanns*
* John von Neumann Institut für Computing, Jülich, Germany
New insights into cellular processes require the synthesis of
information obtained with low- to medium-resolution biophysical
techniques, such as electron microscopy, with atomic-resolution
structures. We are developing novel approaches for the registration of
atomic-resolution subunits with low-resolution densities of large
macromolecular aggregates.
FOURIER TEMPLATE CONVOLUTION
Template convolution takes advantage of Fourier correlation
theory to rapidly scan the 6 translational and rotational degrees of
freedom of a probe molecule relative to a (fixed) target density map
(Fig. 1).
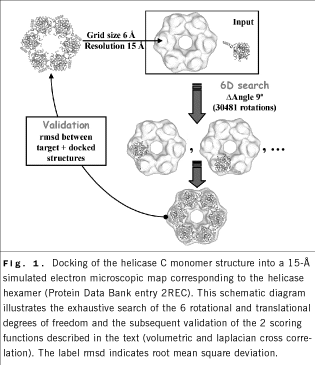 X-ray crystallographic fitting methods, based on volumetric cross correlation, or the R value, are limited to resolutions less than 10 Å, the level at which densities show internal structure.
X-ray crystallographic fitting methods, based on volumetric cross correlation, or the R value, are limited to resolutions less than 10 Å, the level at which densities show internal structure.
The major advantage of our quantitative docking method is that
it extends the practical resolution range to approximately 30 Å by
means of a laplacian operator that emphasizes contour (shape)
information in addition to the traditional volumetric correlation.
Laplacian-filtered density maps maximize the fitting contrast. Tests
with synthetic low-resolution density models of oligomeric assemblies
indicate that the difference in score between the correct fit and
spurious fits increases by 35%-50% relative to the traditional
volumetric correlation-based docking.
FLEXIBLE DOCKING
In the presence of induced-fit conformational changes, rigid-body
docking alone gives poor alignment of the crystal and low resolution.
Therefore, we devised alternative modeling techniques based on
molecular dynamics simulation and point "landmarks" that bring
deviating global features of biopolymers into register while preserving
the crystal structure locally. We added a flexible alignment tool to
our Situs docking package that is based on 3-dimensional "motion
capture" technology used in the entertainment industry and in
biomechanics. By freezing inessential degrees of freedom, skeletons of
connected landmarks significantly reduce the effect of noise and
thereby improve the stereochemical quality of the fitted structures
relative to the unconstrained alignments. In collaboration with E.H.
Egelman, University of Virginia, Charlottesville, we used
skeleton-based flexible fitting to uniquely define a molecular model of
cofilin bound to F-actin that was based solely on the monomeric crystal
structures and on an electron microscopic reconstruction of actin
filaments decorated with human cofilin.
VIRTUAL REALITY
In collaboration with S. Birmanns, John von Neumann Institut für
Computing, Jülich, Germany, we are developing a 3-dimensional graphics
extension for Situs, termed SenSitus, that can support virtual-reality
devices such as stereo glasses, 3-dimensional trackers, and
force-feedback (haptic) devices (Fig. 2).
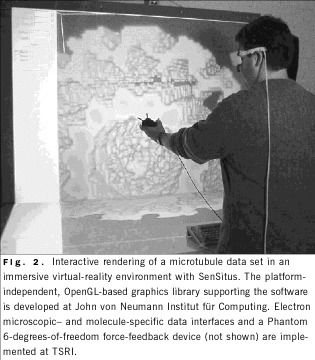 In addition to using automated fitting, microscopists must evaluate and
manipulate docking models interactively "by eye." Three-dimensional
capabilities and the "physics of touch" offer tangible benefits for
modelers who wish to explore a variety of docking situations in a
virtual-reality environment.
In addition to using automated fitting, microscopists must evaluate and
manipulate docking models interactively "by eye." Three-dimensional
capabilities and the "physics of touch" offer tangible benefits for
modelers who wish to explore a variety of docking situations in a
virtual-reality environment.
PUBLICATIONS
Galkin, V.E., Orlova, A., Lukoyanova, N., Wriggers, W., Egelman, E.H.
Actin depolymerizing factor stabilizes an existing state of F-actin and
can change the tilt of F-actin subunits. J. Cell Biol. 153:75, 2001.
Wriggers, W., Birmanns, S. Using Situs for flexible and rigid-body fitting of multi-resolution single molecule data. J. Struct. Biol., in press.
Wriggers, W., Chacón, P. Using Situs for the registration of
protein structures with low-resolution bead models from x-ray solution
scattering. J. Appl. Crystallogr., in press.
Xing, J., Wriggers, W., Jefferson, G.M., Stein, R., Cheung, H.C., Rosenfeld, S.S. Kinesin has three nucleotide-dependent conformations: Implications for strain-dependent release. J. Biol. Chem. 275:35413, 2000.
Wriggers Website
|
|



